About
Organised by CCP5 and sponsored by CECAM, the School is intended for newcomers to the science of molecular simulation and will provide a comprehensive introduction to the theoretical background as well as practical sessions on computational methods and research seminars to illustrate the versatility of simulation in modern research. There will also be opportunities for participants to present their own research.
The Summer School starts with a two-day programming course, where students can opt to take either Python or modern Fortran. After this preparation, the first five days of the main School will cover the basics of molecular simulation, and the remaining three days will be devoted to more advanced courses with options in mesoscale, ab initio, machine learning for interatomic potentials and biomolecular simulation. Course notes will be provided in electronic format. In addition to the lectures, there will be extensive practical sessions in which students will undertake computational exercises to reinforce and further explore the material.
The school will take place between 16th and 27th of July 2023 at Durham University.
A fee of £650 to cover part of the expenses will be charged to successful applicants. The school has 70 places available.
The fee partially covers accommodation, breakfast, lunch and dinner for the duration of the school. It also covers school gala dinner and poster session refreshments. The rest is covered by our sponsors. Successful candidates will need to cover their transport costs.
Please note The school can be recognised towards your doctoral training in UK, also upon request we can provide a letter for ECTS credits for your school.
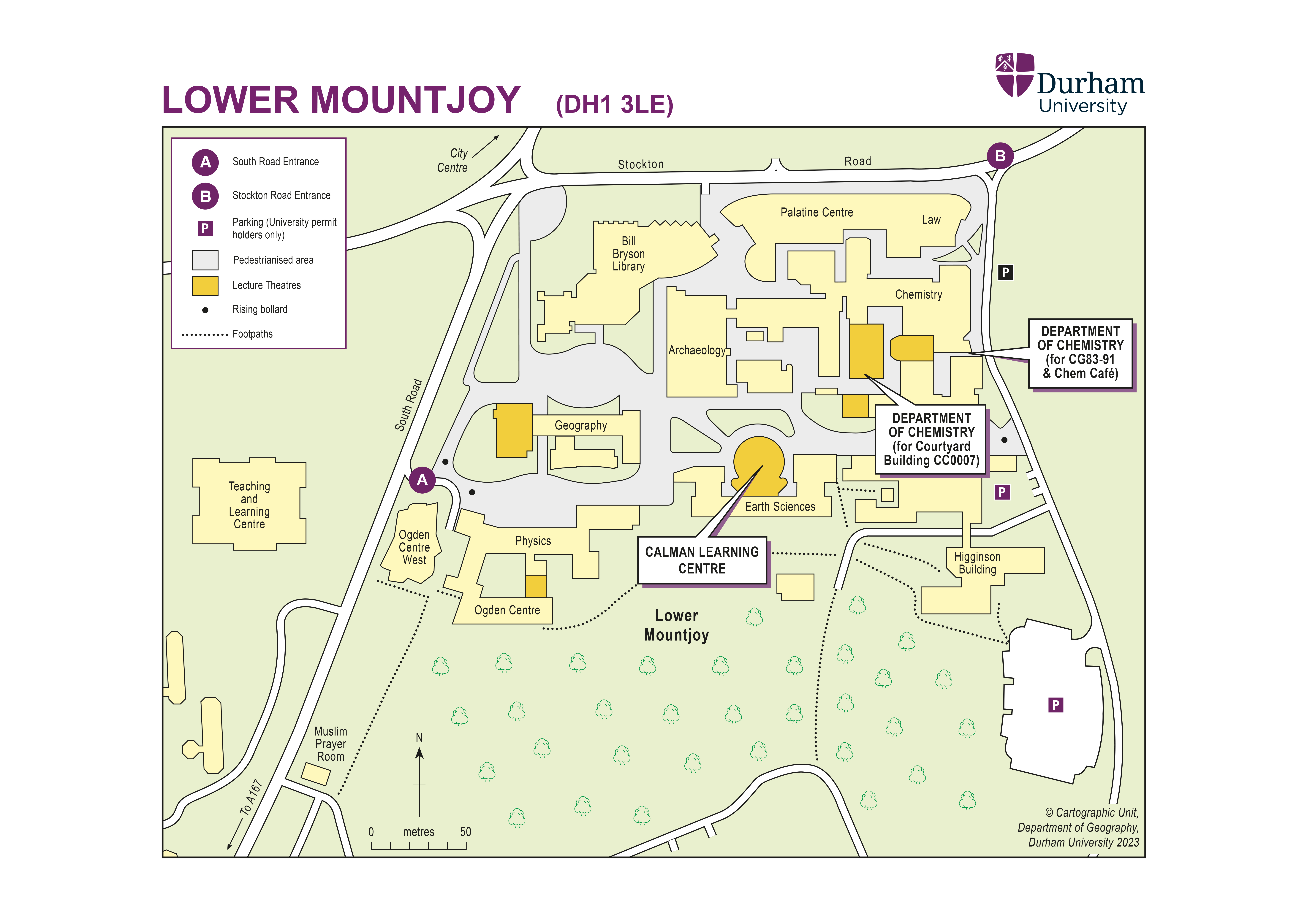
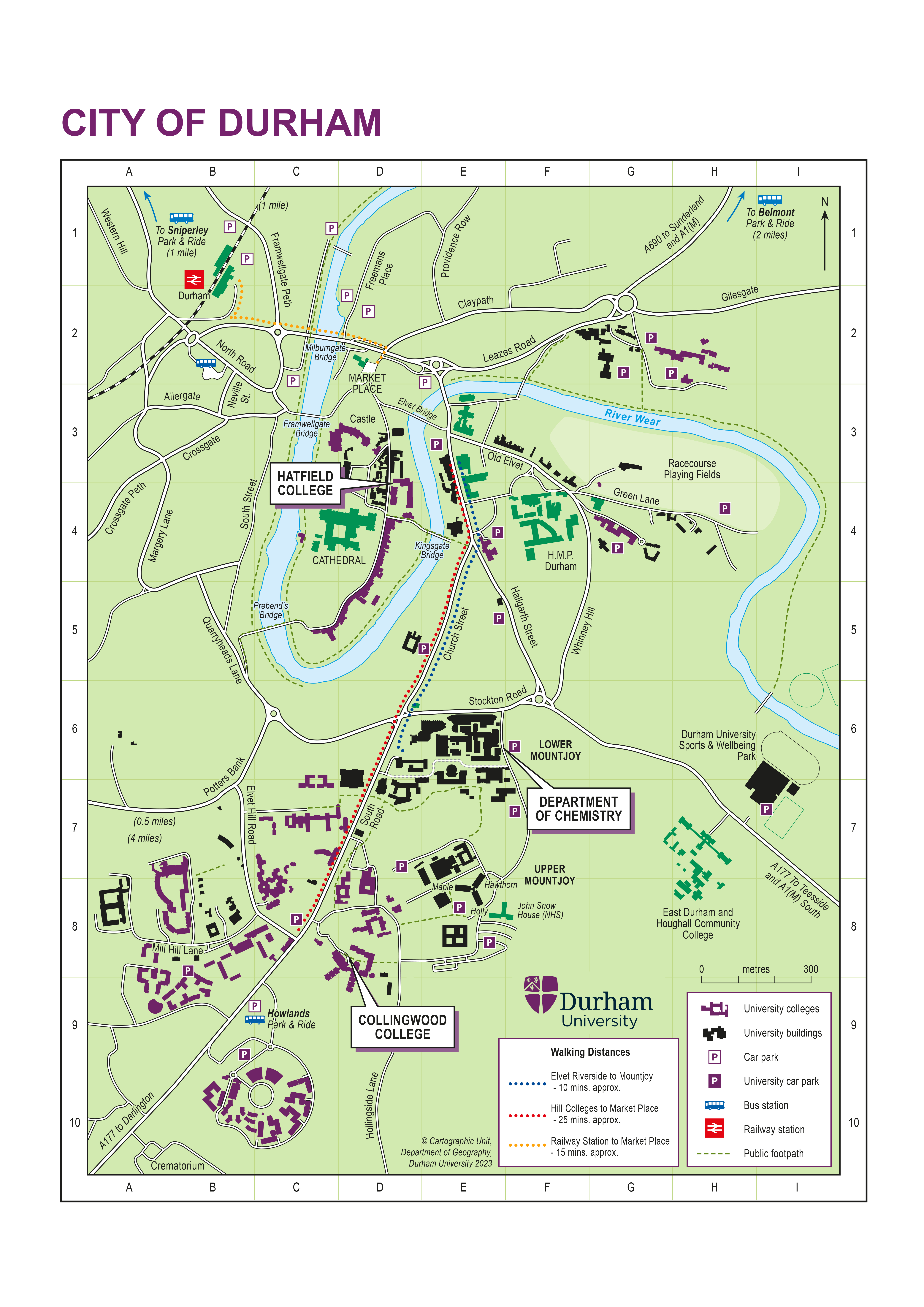
Accommodation will be provided in student accommodation, single en-suite rooms for the duration of the school at Collingwood College, South Road, Durham, DH1 3LT. If you need extra days of accommodation please let us know as soon as possible and you will have to pay for them.
NOTE Initial emails for acceptance have been send… please confirm your place by 10th of April(no need to pay at this stage). Unconfirmed places will go to the waiting list. Please check your SPAM/JUNK folder. If you have not received an email at this stage you are probably on the waiting list.
Key dates
- Application deadline: 15th of March 2023
- Acceptance decision: 31st of March 2023
- Accept place: 10th of April 2023
- Fee payments: 1st of May 2023
Join CCP5

Organising Committee
- Dr Colin Freeman, University of Sheffield
- Prof Neil Allan, University of Bristol
- Dr Mark Miller, University of Durham
- Dr Alin Elena, STFC Daresbury Laboratory
Sponsors






Code of Conduct
We value the participation of everyone and want to ensure that everyone has an enjoyable and fulfilling experience, both professionally and personally. Accordingly, all participants of the CCP5 Summer School are expected to always show respect and courtesy to others. The CCP5 and its partners strive to maintain inclusivity in all of our activities. All participants (staff and students) are entitled to a harassment-free experience, regardless of gender identity and expression, sexual orientation, disability, physical appearance, body size, race, age, and/or religion. Harassment in any form is not acceptable for any of us.
We respectfully ask all attendees of the CCP5 Summer School to kindly conform to the following Code of Conduct:
- Treat all individuals with courtesy and respect.
- Be kind to others and do not insult or put down other members.
- Behave professionally. Remember that harassment and sexist, racist, or exclusionary jokes are not appropriate.
- Harassment includes, but is not limited to, offensive verbal comments related to gender, sexual orientation, disability, physical appearance, body size, race, religion, sexual images in public spaces, deliberate intimidation, stalking, following, harassing photography or recording, sustained disruption of discussions, and unwelcome sexual attention.
- Participants asked to stop any harassing behaviour are expected to comply immediately.
- Contribute to communications with a constructive, positive approach.
- Be mindful of talking over others during presentations and discussion and be willing to hear out the ideas of others.
- All communication should be appropriate for a professional audience, and be considerate of people from different cultural backgrounds. Sexual language and imagery are not appropriate at any time.
- Challenge behaviour, action and words that do not support the promotion of equality and diversity.
- Arrive at the events punctually where possible.
- Show consideration for the welfare of your friends and peers and, if appropriate, provide advice on seeking help.
- Seek help for yourself when you need it.
please report any issues to alin-marin.elena@stfc.ac.uk